|
|
|
|
|
|
| Finding the origin is pretty important to predict the path of the virus in the past and for future. From the history of pathogen outbreaks, question about origin showed many possibilities range from genetics to human errors like spillover, mutations, adaptations, continuous contact with the potential animals, improper handling of organisms in the lab/hospitals/public places etc. |
| In the current scenario many theories have come to link the origin of nCOV-19. Thanks to the developments in NGS technology, which enabled the sequencing of this virus from various sources, in a short span of time. These sequences are made accessible for further analysis through open access databases such as GeneBank, NextStrain, ViPR and GISAID. From the time of first complete sequence availability from Wuhan based study, many interpretations and analyses were published stating the virus roots. If we look into those, one scientist proposed snakes as the possible intermediate host based on the preferential codon usage but the statement couldn’t stand well. The statement saying the virus was from a research institute, also knocked down by a study from the sequencing of environmental samples from the Wuhan seafood market. |
| According to the study of Next Generation Sequencing data of SARS-CoV-2 whole genome, isolated from first set of reported individuals from china, there is >88% similarity at nucleotide level with Bat SARS like coronavirus (Bat SL CoV) than SARS CoV (79% homology) and MERS CoV ( 50% homology). This derives a conclusion that the new virus has more close ancestry with bat viruses but not with SARS and MERS virus. One more apparent observation that was showed is, similarity of >99.9 % among the new sequences. This definitely say that the virus origin was from bats or both might have had a common ancestor. However we don’t have the evidence in terms of sequence to say that, the virus has spilled over directly from bats or through an intermediate species. If we see the source virus sequences from previous out breaks, there is only a difference of 10 nucleotides or less between SARS virus and Civet cat virus, MERS virus and Dromedary Camel virus. So lack of such kind of strong similarity with existing sequences is creating a gap to say how and when the virus infected humans first. |
| Though there is a great level of homology with Bat viruses, the overall genome structure is similar to SARS CoV as you can see from picture 3. From the study of J F-W Chan et al, there are many regions of spike protein showing similarity with SARS-CoV. If we see the alignment data for spike protein there is a homology with SARS CoV than Bat CoV. In Bat CoV genome there is a stretch of missing amino acids at two places in the receptor binding domain. Those parts in the SARS CoV and SARS CoV-2 are having amino acids that are showing slight similarity. That might be the reason behind the successful ACE-2 receptor binding for human invasion. As showed in the picture below there is a difference in the size of the spike protein as well. |
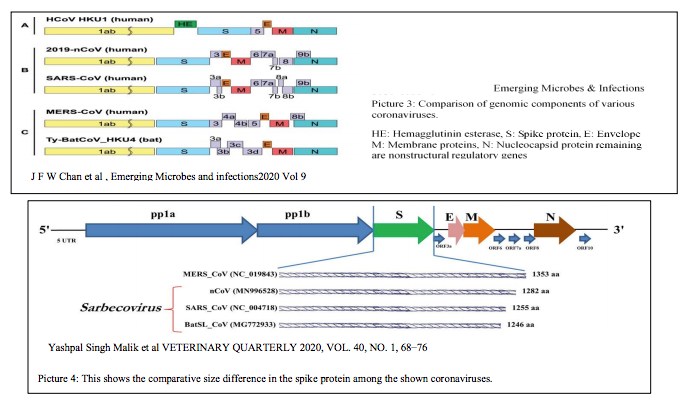
|
| Another analysis by Bedford says that, another bat virus RaTG13 is having more sequence similarity (96%) with current virus with a difference of only 1100 nucleotides. This sequence is not isolated from the direct virus but from the fecal sample of a bat in Yunnan region. From the known coronavirus mutation rate, he is stating both viruses might have shared a common ancestor 25 to 65 years ago. |
|
|
| SARS-CoV-2 mainly attacks the epithelium cells in the secretory tissues such as eyes, nose and the alveoli cells through ACE-2 receptors same like SARS virus. But there is a potential possibility of affecting other organs such as small intestine, colon, and kidney due to the prevalence in the expression of ACE 2 receptors. However, few clinical study reports showed the ability of these viruses in crossing the blood brain barrier where no ACE2 expression and no invasion in the tissues where there is substantial receptor expression indicates a need for further study in this Cellular entry mechanism. |
| There is a 72% homology in the RBD of SARS CoV and SARS CoV 2. However symptoms are related to lower respiratory system such as dry cough, sputum, dyspnea, ARDS-pneumonia but varies from person to person. The active replication and macrophage interaction happens at type-2 pneumocytes in the alveoli. This eventually alerts immune system by producing interleukins, interferons, and other cytokines. In 85% of the studied cases, the symptoms were mild flu like such as fever, myalgia and cough but subside in 7-8 days if taken care in terms of body hydration, rest etc. to boost immunity. In other cases it showed more severe kind of immune response that actually manifests in symptoms such as pneumonia, ARDS that finally led to death when coupled with other superinfections and preexisting illnesses. |
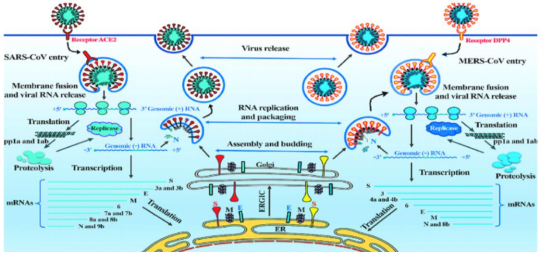
|
| Zhiqi Song, Yanfeng Xu, Linlin Bao et.al, Viruses 2019, 11, 59 Picture 6: Shows the complete picture from virus entry into to release from host cell. SARS-CoV uses ACE 2 receptor while MERS CoV uses DPP 4 or CD 4 receptor on the target cell to enter |
| Initially they thought infection was confined among family clusters and healthcare workers. But this virus took a quick hop in holding many countries across globe. This can impart to the R naught value which is relatively more for this novel virus than SARS and MERS viruses. The study of Chen et al, says that the differences like having a flexible glycyl residues in the distinct loop instead of prolines and presence of phenyl alanine F486 in the flexible loop might have pushed for the stronger interactions between RBD and hydrophobic pocket of ACE2 receptor. In addition to that, the infected persons with mildest symptoms, could still transmit the virus to healthy people. Unlike the flu virus its transmission most likely doesn’t change by climatic conditions. On top of the above facts, lack of approved medication and vaccination made it difficult to contain. |
| Till now the most popular route of human to human transmission happened through contact with sneeze, breathe and cough droplets up to a distance of at least 3 meters, and through surfaces or appliances that has virus. There is also evidence that the virus was even detected in feces of affected individuals along with blood, sputum and other upper and lower respiratory fluids. From the disease statist’s observation of past cases, men were more among infected than women and children and adolescents were least affected. It doesn’t transmit to womb through placenta in pregnant women and doesn’t transmit through lactation, from a study report. However a study from Johns Hopkins Bloomberg School of Public Health concluded that children are equally susceptible to this virus infection and is same like any other adults when exposed to the virus. |
|
|
RT-qPCR/multiplex PCR: This test method is suitable for samples such as nasal swab, anal swab, lung fluids and blood from affected individual. The real time primers that target part of the S gene or ORF 1ab region to confirm. The test takes 4 to 5 hours to complete and requires special facility and a person with technical skills to handle the test. |
|
iLACO colorimetric detection: This is isothermal LAmp based method for COvid-19 detection a newly adapted technique. This method also based on amplification of a specific region of the genome and can be carried out in a simple temperature regulated water bath. This test uses 3 sets of specific primers for a region of ORF 1ab, viral RNA template that was purified from swab or blood and other detection reagents. When incubated at 65°C for 20 min in a water bath, positive test gives a yellow color reaction and negative test remains pink. |
|
ELISA: Serum from effected persons can be analyzed for novel virus specific IgG, IgM antibody levels using antigen Rp3 Nucleocapsid protein from SARSr-CoV ELISA kits. |
|
Chest X ray/CT: In severe cases with lung lesions show ground glass appearance in X-ray and CT films. |
|
| From the time the virus started spreading to the countries, many scientists from institutes and organizations are working hard, in finding methods to treat and prevent this coronavirus infection through new antibodies, drugs and vaccines. |
| Spike protein is a proven target which is on the surface of the virus. It is a trimer with receptor binding domain (RBD) at the active site. Successful interaction of this domain with ACE-2 receptors is key to the host cell invasion. This part of the virus sequence is one of the conserved regions of the genome which determines host specificity and range. Neutralizing antibodies against the spike protein RBD can be an excellent option as many such sites are exposed on the virus surface to make strong complex rendering its further entry. For successful entry of this virus in to host cell require another process called spike priming, which is done by a host serine protease named TMPRSS-2. Targeting this protein with an inhibitor camostat mesylate, showed inhibition of virus entry into the primary human lung cells and calu-3 cell lines. This gives a second option in hand when the first one fails. These sort of drugs in combination with immune boosting therapeutics such as monoclonal antibody therapy or interferon therapy might play effectively to curb the infection severity. |
| Just one or two proteins or receptor compatibility cannot break such a huge and amazing system of humans. As shown in the below picture, there should be some more rate limiting steps used by virus to decode the host specificity and successful invasion. A lot more research is required to study the each step of the virus in the host. Such information might be useful to develop broad spectrum antivirals for future. The following preexisting drugs that were proven effective, in the past times are now under reevaluation to test against SARS-CoV-2. |
 Showing possible targets for the future therapeutics from the study of Xiaohuan Wang, Peng Zou et.al
Showing possible targets for the future therapeutics from the study of Xiaohuan Wang, Peng Zou et.al
|
| Remdesivir: This is a nucleotide analog and used effectively during Ebola, SARS and MERS out breaks. There is a report that this was used in combination with interferon, in Wuhan as well to treat hospitalized patients. This drug showed positive effect in in vitro studies against SARS-CoV-2. |
| Oseltamivir: This is a neuraminidase inhibitor that prevents the release of viral progeny from host cells. There is no enough data supporting its use. |
| Lopinavir: This is an anti-retroviral protease inhibitor used in HIV treatment however showed no apparent benefit against novel coronavirus according to a study. |
| Hydroxychloroquine: This drug showed effect in invitro studies against SARS-CoV-2. A France based study showed some positive outcome upon use of a malaria drug named hydroxychloroquine combined with azithromycin. Another study from china said, this drug reportedly reduced the hospital stay and recovery time in studied patients. Considering the positive study outcomes it got approval from FDA for its further trials. A supportive study in cell lines by Jia Liu et al showed the possible mechanism of action of HCQ in the COVID-19 scenario. One is that HCQ being a weak base, has increased the endosomal pH from 4.5 to 6.5 preventing the vesicle maturation. This successfully inhibit the virion release from cell. In addition, due to its proven anti-inflammatory effect, it can markedly reduce the production of cytokines to alleviate the intensity of pneumonia and ARDS. |
| An USA based company Moderna Inc., specialized in developing RNA/mRNA based therapeutics and vaccines for various illnesses, started its phase I trial for COVID-19 vaccine mRNA-1273. Rocky Mountain laboratories in collaboration with Oxford University is in the development of chimpanzee adenovirus-vector based vaccine for COVID-19. A group of scientists are trying to check the efficacy of SARS vaccine against COVID-19. |
| This spike protein is a trimeric protein which shows confirmation change represented as Pre-fusion to Post-fusion state upon binding to the receptor. A team from University of Queensland developed a vaccine candidate to mimic the pre fusion form using molecular clamp technology. As the pre-fusion form of spike protein has the more chance to be recognized by our immune system they are expecting this can be a better vaccine candidate. This vaccine is ready to be tested in animals. |
| There is another vaccine ready for testing from Novavax. This vaccine also based on Spike protein but in combination with company’s proprietary Matrix-M™ adjuvant developed through recombinant nanoparticle technology platform. |
| Based on their comparative study between IBV and SARS-CoV-2 a biotechnology team led by Dr. Chen Katz from MIGAL, is confidant on its attempt to customize IBV (poultry virus) vaccine to make it work for the novel corona virus. According to Zeng Yixin from China’s National Health Commission, they are trying in five different approaches including inactivated whole virus, engineered antigens, modifying influenza vaccine etc. to come up with an effective solution. |
|
| This novel virus has low fatality rate but more infectivity rate than its peers which is alarming to call for quick containment. The reason behind this is being RNA virus it can make populations of quasispecies through mutations and homologous recombination. The more it replicates in a species, the more it can accumulate mutations that can generate new strains because of longer selection pressure. So quick containment is very crucial before its genome encounter any more potential chances to make it more virulent. |
| In this scenario Self-isolation is considered as a best way to cut the transmissibility cascade among people. There is a mathematical simulation that was published recently, explaining the importance of selfisolation to combat this pandemic. This slowdown in the virus transmission is actually advantageous as it gives time to select less virulent long living groups in the circulating populations through attenuation. In addition to that, it gives the health care professionals ample time, resources and other supporting facilities to treat severely affected persons. |
| Personal hygiene practices such as washing hands with soap for at least 20s, disinfecting with any liquid having at least 60% ethyl alcohol, proper disposing of used masks and other biological wastes from infected individuals are like physical barriers for virus entry along with quarantine measures. |
|
| At this stage, it is good that we shut down operations, self-limiting ourselves to homes to break the chain of Virus spread. It is also true that our governments and hospitals are working hard to give greatest available service by facilitating more clinics and hospitals to serve the affected, along with educating and enforcing the necessary steps if required. |
| At this point we are not certain about its containment, as there is also a possibility that it can remain as an endemic to a particular region or country, according to some experts. However everyone is pretty much aware that, soon many of the things comes to normal. No matter how well we handled the situation this time, we have no clue who is going to be our next enemy? So if we don’t update our technology at the pace of virus updating its genome, we have to go through situations same like this or even worse? in future. The governments should encourage more and more research in this line. Unless we have handful of strategies for prevention & control through sustainable technologies in terms of general disinfection, sterilization and broad spectrum antivirals & anti-bacterials it is going to be a disaster. Because we don’t know how strong our next pathogen is? |
| Despite the advancement in terms of scientific technology, still needs more inputs from the experts to produce quick in hand screening methods and broad spectrum therapeutic options to face unforeseen times in the future. Along with this if researchers can really put efforts EQUALLY on all the strategies such as vaccinations, quick economical screening methods, Safe & wide number of Sanitization & sterilization methods, broad spectrum anti-microbial therapeutics, and alternative medicine methods, it will be easy to execute quick action plans according to the need. |
| Not having complete information or data in particular to pathogen may hamper the treatment, but why are we restricting ourselves from executing regulated disinfection & sterilization practices in any country to combat this highly infectious virus? In everyday life Sanitation, disinfection and sterilization methods are basic strategies that are saving the mankind from many microscopic pathogens. Research in this can be fruitful in giving more promising options that can be used for wide range of pathogens. This might matter in the countries that require such kind of standards because even a small outbreak has the potential to spread to the world. |
| It is a known fact in Microbiology that Viruses can be easily denatured by many disinfection methods. Yes, 70% alcohol is effective in deactivating microbial activity, but we should not rely on one methodology to wipe out this virus from surfaces. And also this virus can stay up to 3 hours in Air. Besides treating the patients, simultaneous, deactivation of the activity of the virus (usually made up of lipoprotein components with DNA/RNA) in atmosphere or on any surfaces, using available sterilization method or disinfection methods is important. |
| Hydrogen peroxide at diluted strengths works as an excellent disinfectant and relatively safe than some of the chemicals of this kind. Center for disease control has already listed its potential antimicrobial activity and sporicidal activity. Hydrogen peroxide is one of those and safe to use in hospital settings to sanitize devices, air and to disinfect hazardous biological samples/waste. Besides that, in alternative medicine there is a practice of using diluted hydrogen peroxide mist to treat patients with various respiratory issues including asthma and COPD. There is some supporting evidences, that showed promising results when used hydrogen peroxide therapy for respiratory illnesses. Further research in this line might open new options for clinical treatments and sanitization methods based on this alone or in combination with other compatible chemicals. |
| If we could find solutions through innovative & sustainable technology it can really save the world. For example developing masks that are reusable and masks that are having bactericidal/virucidal/sporicidal properties. This will be useful if any pathogen is airborne or transmit through aerosols. There is already wide range of information available from past research about such microbicide agents. It would be great if such technology reach to people, where PPE not only acts as physical barrier but also can kill the pathogen when it encounters. Doesn’t it sound amazing? |
| While researching the corona virus pathology in general, came across couple of research papers showing their capability to cross blood brain barrier. There is a recent report of death due to MERS with clinical symptoms affecting brain. This is alarming and also giving a hint that there is a possibility that humans may contact new versions of this or for that matter any microbe with more pathogenicity and infectivity in future. If we see the previous out breaks, all viruses has link to bats at some point of their evolution. The period of 15 years in the evolution generated three new successful versions of a virus that can infect humans from non-human virus. But that same experience and time couldn’t really generate promising therapeutics that can be used in the present scenario. Change in the direction of our research would be of any help? By understanding the immune system of bats along with its genome might give valuable clues for future solutions. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|




